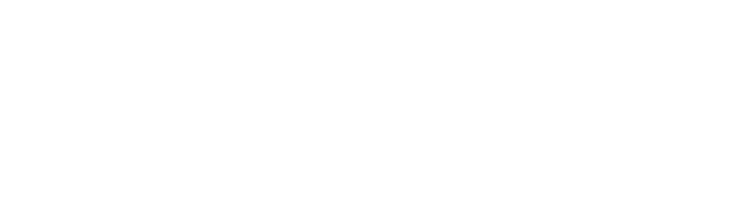Dr. Charles Wang's current research focuses on substance use disorders using prenatal exposure animal models to understand the genomic and epigenomic mechanisms underlying maternal drug exposure-induced abnormal brain development.
Genomic and epigenomic mechanisms of maternal e-cigarette (e-cig) smoking-induced abnormal brain development
The overall goal of the study is to investigate the genomic and epigenomic mechanisms underlying abnormal brain development caused by prenatal e-cigarette exposure. Maternal smoking is a well-recognized public health concern associated with increased neurodevelopmental disorders and other health risks in offspring. Substantial evidence indicates various adverse effects of maternal smoking or prenatal nicotine exposure on neonatal brain development, e.g., hyperactivity, reduced cognitive performance, increased anxiety and depression, and increased susceptibility to brain injury. Growing evidence suggests that maternal e-cig use affects brain development, resulting in abnormal cortical neuronal morphology and aberrant neuro-behaviors in offspring.
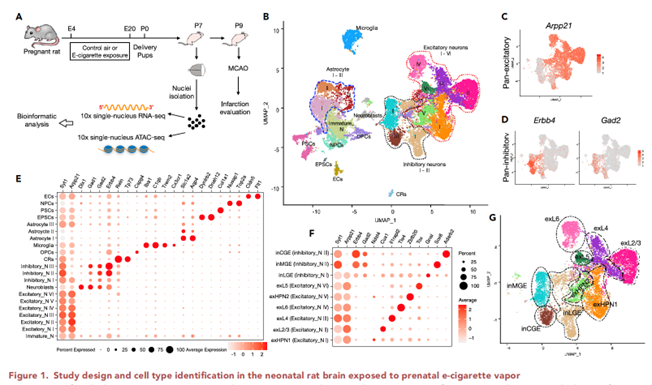
Using a rat prenatal e-cig exposure model and single-nucleus sequencing, we recently found that prenatal e-cig exposure disrupted excitatory-inhibitory (E/I) balance, ratio of excitatory/ inhibitory neurons, in the neonatal brain. E/I balance is crucial for normal brain development and functions, and its disruption has been postulated to underlie the pathogenesis of many neurodevelopmental disorders, including autism spectrum disorders, ADHD, and other psychiatric disorders. However, the underlying genomic and epigenomic mechanisms are still not known. This investigation seeks to fill this knowledge gap.
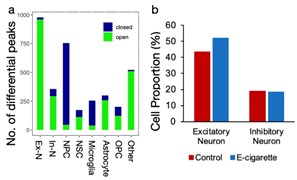
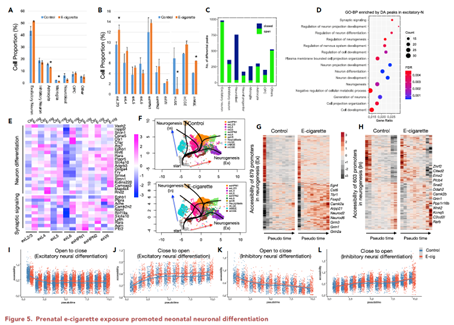
Our hypothesis is that prenatal e-cig exposure induces epigenomic reprogramming and genomic alterations in the developing brain, which cause an E/I imbalance and consequently an increased risk for neurodevelopmental disorders. Exploiting the prenatal e-cig exposure model developed, we intend to achieve three objectives. Aim 1 is to determine the spatial-temporal effects of prenatal e-cig exposure on brain development and progression of E/I imbalance using spatial genomics and single-cell sequencing techniques. Aim 2 is to determine the epigenomic mechanisms regulating the prenatal e-cig induced E/I imbalance. Aim 3 is to investigate whether prenatal e-cig causes genomic alterations (i.e., SNVs and CNVs) that are involved in E/I imbalance. This project is funded by NIH under the NIDA Animal Genomics Consortium. Many cutting-edge genomic and epigenomic technologies are exploited in our studies, including spatial genomics, single-nucleus RNA-seq and chromatin accessibility (snATAC-seq), and state-of-the-art bioinformatics tools. We expect that this study will provide valuable new insights into the effects of e-cig on the early central nervous system development, which will help explore promising molecular and cellular therapeutic targets for treating e-cig vaping-induced brain damage.
Investigating the underlying epigenomic mechanisms of prenatal Cannabis (THC) exposure-induced abnormal brain development:
The overall goal of the study is to investigate the effects of prenatal THC exposure induced abnormal brain development and the underlying epigenomic mechanisms. Marijuana (Cannabis) is one of the most widely used psychotropic drugs in the United States. Cannabis use among pregnant women in the US is increasing with prevalence as high as 14% among 12–18-year-old pregnant women. Accumulating evidence has clearly demonstrated that prenatal exposure to D9- tetrahydrocannabinol (THC), the major psychoactive component of cannabis, has long-term negative effects on decision- making and molecular disturbances linked to synaptic plasticity with profound epigenetic dysregulations that are exacerbated by stress. However, the underlying genomic and epigenomic pathophysiologic mechanisms are still not known. In this study, we investigate whether prenatal THC exposure will lead to altered behavior in rat offspring and impact the intricate connections of neural circuits which may be orchestrated by epigenomic reprogramming in the developing rat brain. Many cutting-edge genomic and epigenomic technologies are exploited in this study, including spatial transcriptomics, single-nucleus RNA-seq and chromatin accessibility (snATAC-seq), and state-of-the-art bioinformatics tools.
There are many other projects currently supported by Loma Linda University School of Medicine's Center for Genomics involving in genomics, epigenomics, single-cell sequencing and bioinformatics in collaborating with many different investigators at Loma Linda University, including cancer genomics and clinical genomics.
Other Publications
Featured Publications
Precision Clinical Medicine I March 2023
Liu T, Chen Z, Chen W, Evans R, Xu J, Reeves ME, de Vera ME, Wang C. Dysregulated miRNAs modulate tumor microenvironment associated signaling networks in pancreatic ductal adenocarcinoma. doi: 10.1093/pcmedi/pbad004. PMID: 37007745; PMCID: PMC10052370.
Precision Clinical Medicine 2023 Mar 10;6(1):pbad004
Communications Biology I April 2023
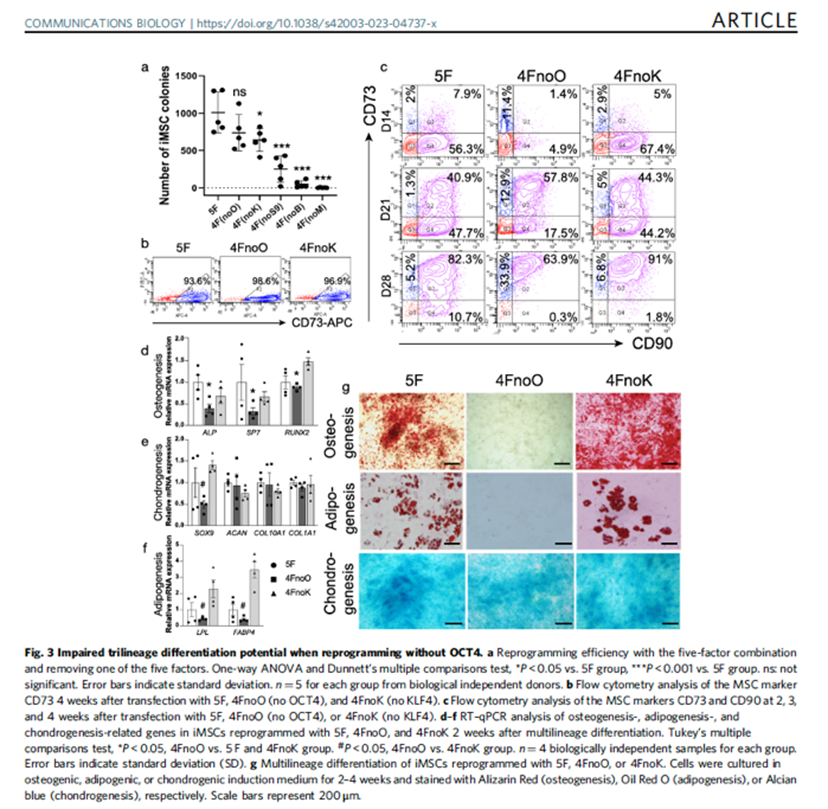
Chen W, Wang C, Yang ZX, Zhang F, Wen W, Schaniel C, Mi X, Bock M, Zhang XB, Qiu H, Wang C (corresponding). Reprogramming of human peripheral blood mononuclear cells into induced mesenchymal stromal cells using non-integrating vectors. doi: 10.1038/s42003-023-04737-x. PMID: 37041280; PMCID: PMC10090171.
Communications Biology 2023 Apr 11;6(1):393
iScience I June 2022
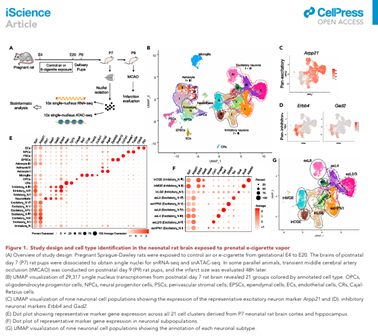
Chen Z, Chen W, Li Y, Moos M Jr, Xiao D, Wang C (corresponding). Single-nucleus chromatin accessibility and RNA sequencing reveal impaired brain development in prenatally e-cigarette exposed neonatal rats. doi: 10.1016/j.isci.2022.104686. PMID: 35874099; PMCID: PMC9304611.
iScience 2022 Jun 30;25(8):104686
Gut Microbes I January 2022
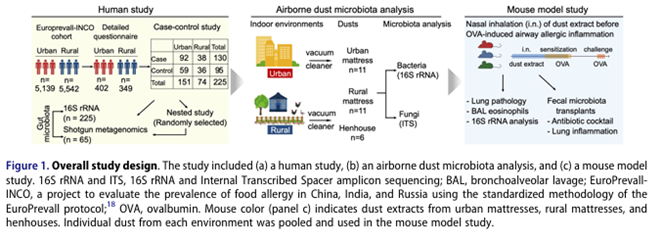
Yang Z, Chen Z, Lin X, Yao S, Xian M, Ning X, Fu W, Jiang M, Li N, Xiao X, Feng M, Lian Z, Yang W, Ren X, Zheng Z, Zhao J, Wei N, Lu W, Roponen M, Schaub B, Wong GWK, Su Z, Wang C (co-corresponding), Li J. Rural environment reduces allergic inflammation by modulating the gut microbiota. doi: 10.1080/19490976.2022.2125733. PMID: 36193874; PMCID: PMC9542937. 122
Gut Microbes 2022 Jan-Dec;14(1):2125733

Genome Biology I November 2022
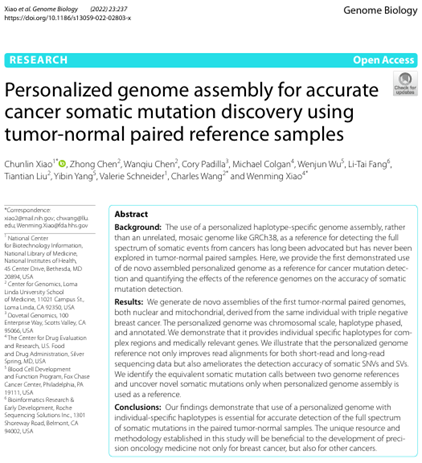
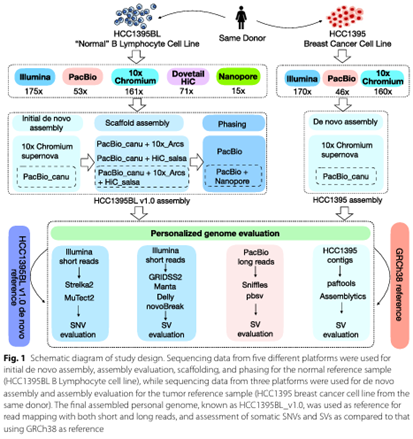
Xiao C, Chen Z, Chen W, Padilla C, Colgan M, Wu W, Fang LT, Liu T, Yang Y, Schneider V, Wang C (co-corresponding), Xiao W. Personalized genome assembly for accurate cancer somatic mutation discovery using tumor-normal paired reference samples. doi: 10.1186/s13059-022-02803-x. PMID: 36352452; PMCID: PMC9648002.
Genome Biology 2022 Nov 9;23(1):237
Genome Biology I December 2022
Talsania K, Shen TW, Chen X, Jaeger E, Li Z, Chen Z, Chen W, Tran B, Kusko R, Wang L, Pang AWC, Yang Z, Choudhari S, Colgan M, Fang LT, Carroll A, Shetty J, Kriga Y, German O, Smirnova T, Liu T, Li J, Kellman B, Hong K, Hastie AR, Natarajan A, Moshrefi A, Granat A, Truong T, Bombardi R, Mankinen V, Meerzaman D, Mason CE, Collins J, Stahlberg E, Xiao C, Wang C (co-corresponding), Xiao W, Zhao Y. Structural variant analysis of a cancer reference cell line sample using multiple sequencing technologies. doi: 10.1186/s13059-022-02816-6. PMID: 36514120; PMCID: PMC9746098
Precision Clinical Medicine I January 2021
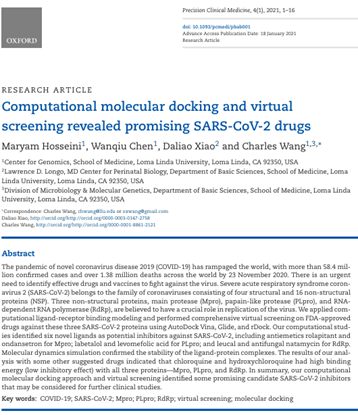
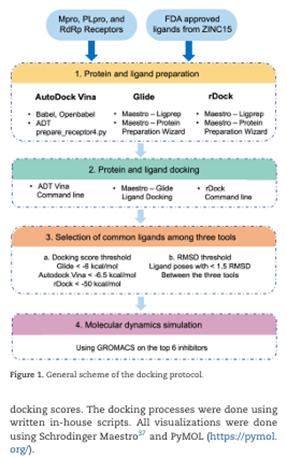
Hosseini M, Chen W, Xiao D, Wang C (corresponding). Computational molecular docking and virtual screening revealed promising SARS-CoV-2 drugs. doi: 10.1093/pcmedi/pbab001. PMID: 33842834; PMCID: PMC7928605.
Precision Clinical Medicine 2021 Jan 18;4(1):1-16
Precision Clinical Medicine I May 2021
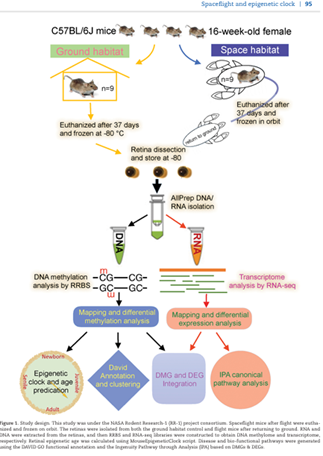
Chen Z, Stanbouly S, Nishiyama NC, Chen X, Delp MD, Qiu H, Mao XW, Wang C (corresponding). Spaceflight decelerates the epigenetic clock orchestrated with a global alteration in DNA methylome and transcriptome in the mouse retina. doi: 10.1093/pcmedi/pbab012. PMID: 34179686; PMCID: PMC8220224.
Precision Clinical Medicine 2021 May 17;4(2):93-108

iScience I August 2021
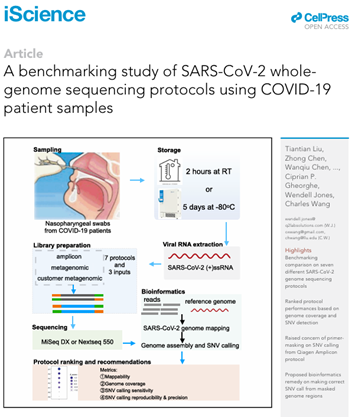
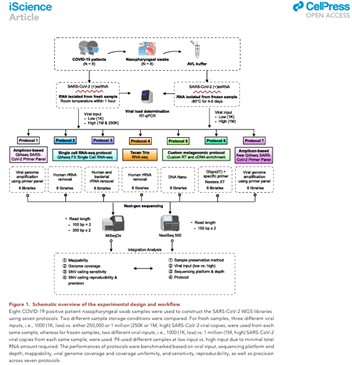
Liu T, Chen Z, Chen W, Chen X, Hosseini M, Yang Z, Li J, Ho D, Turay D, Gheorghe CP, Jones W, Wang C (corresponding). A benchmarking study of SARS-CoV-2 whole-genome sequencing protocols using COVID-19 patient samples. doi: 10.1016/j.isci.2021.102892. Epub 2021 Jul 21. PMID: 34308277; PMCID: PMC8294598.
iScience 2021 Aug 20;24(8):102892
Nature Biotechnology I September 2021
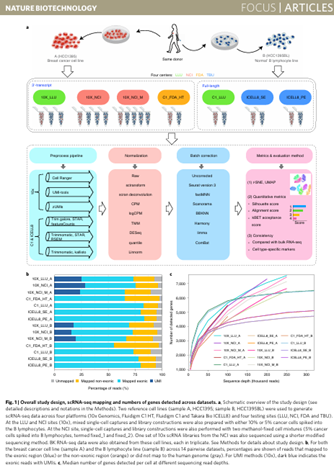
Chen W, Zhao Y, Chen X, Yang Z, Xu X, Bi Y, Chen V, Li J, Choi H, Ernest B, Tran B, Mehta M, Kumar P, Farmer A, Mir A, Mehra UA, Li JL, Moos M Jr, Xiao W, Wang C (corresponding). A multicenter study benchmarking single-cell RNA sequencing technologies using reference samples. doi: 10.1038/s41587-020-00748-9. Epub 2020 Dec 21. PMID: 33349700; PMCID: PMC11245320.
Nature Biotechnology 2021 Sep;39(9):1103-1114
Received the Best Paper Award, at the 2021 MAQC Society Annual Meeting!

Nature Biotechnology I September 2021
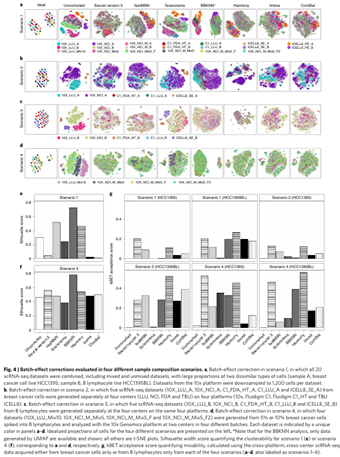
Xiao W, Ren L, Chen Z, Fang LT, Zhao Y, Lack J, Guan M, Zhu B, Jaeger E, Kerrigan L, Blomquist TM, Hung T, Sultan M, Idler K, Lu C, Scherer A, Kusko R, Moos M, Xiao C, Sherry ST, Abaan OD, Chen W, Chen X, Nordlund J, Liljedahl U, Maestro R, Polano M, Drabek J, Vojta P, Kõks S, Reimann E, Madala BS, Mercer T, Miller C, Jacob H, Truong T, Moshrefi A, Natarajan A, Granat A, Schroth GP, Kalamegham R, Peters E, Petitjean V, Walton A, Shen TW, Talsania K, Vera CJ, Langenbach K, de Mars M, Hipp JA, Willey JC, Wang J, Shetty J, Kriga Y, Raziuddin A, Tran B, Zheng Y, Yu Y, Cam M, Jailwala P, Nguyen C, Meerzaman D, Chen Q, Yan C, Ernest B, Mehra U, Jensen RV, Jones W, Li JL, Papas BN, Pirooznia M, Chen YC, Seifuddin F, Li Z, Liu X, Resch W, Wang J, Wu L, Yavas G, Miles C, Ning B, Tong W, Mason CE, Donaldson E, Lababidi S, Staudt LM, Tezak Z, Hong H, Wang C (co-corresponding), Shi L. Toward best practice in cancer mutation detection with whole-genome and whole-exome sequencing. doi: 10.1038/s41587-021-00994-5. Epub 2021 Sep 9. PMID: 34504346; PMCID: PMC8506910
Nature Biotechnology 2021 Sep;39(9):1141-1150


Nature Biotechnology I September 2021
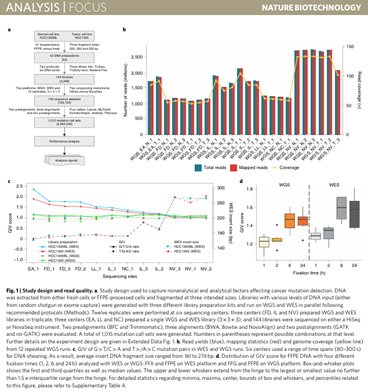
Fang LT, Zhu B, Zhao Y, Chen W, Yang Z, Kerrigan L, Langenbach K, de Mars M, Lu C, Idler K, Jacob H, Zheng Y, Ren L, Yu Y, Jaeger E, Schroth GP, Abaan OD, Talsania K, Lack J, Shen TW, Chen Z, Stanbouly S, Tran B, Shetty J, Kriga Y, Meerzaman D, Nguyen C, Petitjean V, Sultan M, Cam M, Mehta M, Hung T, Peters E, Kalamegham R, Sahraeian SME, Mohiyuddin M, Guo Y, Yao L, Song L, Lam HYK, Drabek J, Vojta P, Maestro R, Gasparotto D, Kõks S, Reimann E, Scherer A, Nordlund J, Liljedahl U, Jensen RV, Pirooznia M, Li Z, Xiao C, Sherry ST, Kusko R, Moos M, Donaldson E, Tezak Z, Ning B, Tong W, Li J, Duerken-Hughes P, Catalanotti C, Maheshwari S, Shuga J, Liang WS, Keats J, Adkins J, Tassone E, Zismann V, McDaniel T, Trent J, Foox J, Butler D, Mason CE, Hong H, Shi L, Wang C (co-corresponding), Xiao W; Somatic Mutation Working Group of Sequencing Quality Control Phase II Consortium. Establishing community reference samples, data and call sets for benchmarking cancer mutation detection using whole-genome sequencing. doi: 10.1038/s41587-021-00993-6. Epub 2021 Sep 9. PMID: 34504347; PMCID: PMC8532138.
Nature Biotechnology 2021 Sep;39(9):1151-1160

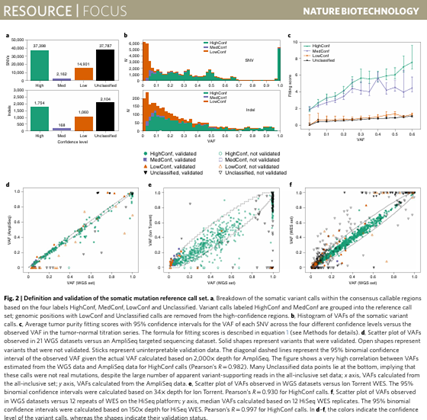
Precision Clinical Medicine I August 2021
Liu MM, Liu T, Yeung S, Wang Z, Andresen B, Parsa C, Orlando R, Zhou B, Wu W, Li X, Zhang Y, Wang C (co-corresponding), Huang Y. Inhibitory activity of medicinal mushroom Ganoderma lucidum on colorectal cancer by attenuating inflammation. doi: 10.1093/pcmedi/pbab023. PMID: 35692861; PMCID: PMC8982591.
Precision Clinical Medicine 2021 Aug 28;4(4):231-245

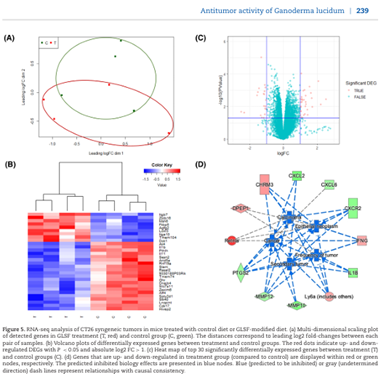
Scientific Reports I October 2020
Zhou N, Chen X, Xi J, Ma B, Leimena C, Stoll S, Qin G, Wang C (co-corresponding), Qiu H. Novel genomic targets of valosin-containing protein in protecting pathological cardiac hypertrophy. doi: 10.1038/s41598-020-75128-z. PMID: 33093614; PMCID: PMC7582185.
Scientific Reports 2020 Oct 22;10(1):18098

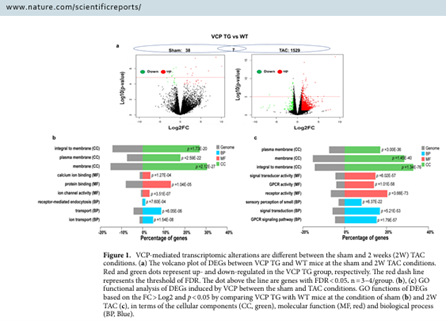
Precision Clinical Medicine I December 2020
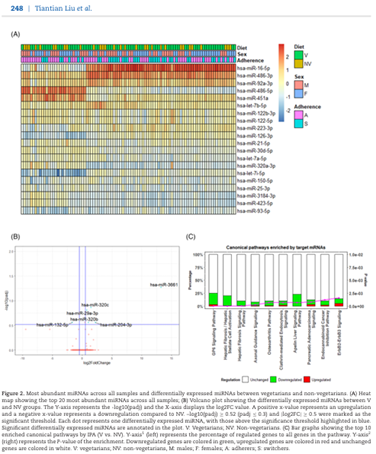
Liu T, Gatto NM, Chen Z, Qiu H, Lee G, Duerksen-Hughes P, Fraser G, Wang C (corresponding). Vegetarian diets, circulating miRNA expression and healthspan in subjects living in the Blue Zone. doi: 10.1093/pcmedi/pbaa037. Epub 2020 Oct 23. PMID: 33391847; PMCID: PMC7757436.
Precision Clinical Medicine 2020 Dec;3(4):245-259

Cardiovascular Research I July 2019
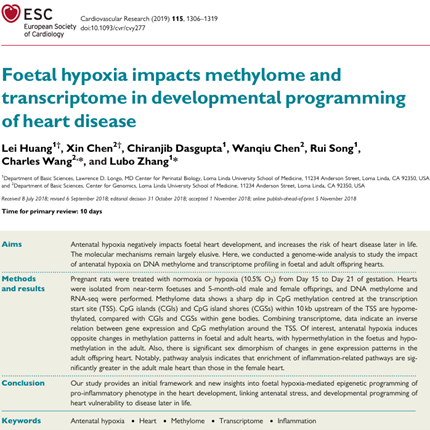
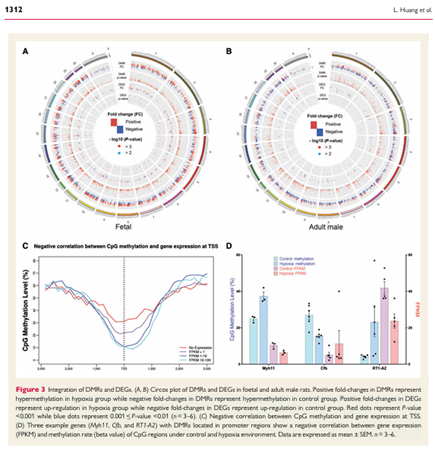
Huang L, Chen X, Dasgupta C, Chen W, Song R, Wang C (co-corresponding), Zhang L. Foetal hypoxia impacts methylome and transcriptome in developmental programming of heart disease. doi: 10.1093/cvr/cvy277. PMID: 30395198; PMCID: PMC6587923.
Cardiovascular Research 2019 Jul 1;115(8):1306-1319.
Precision Clinical Medicine I December 2019
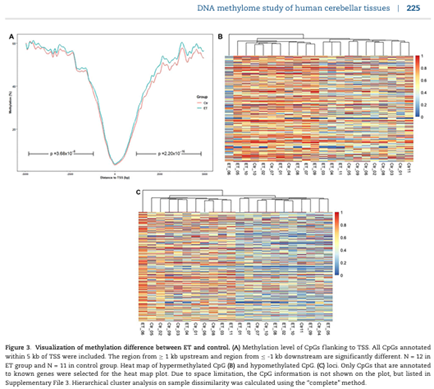
Paul JL, Dashtipour K, Chen Z, Wang C (corresponding). DNA methylome study of human cerebellar tissues identified genes and pathways possibly involved in essential tremor. doi: 10.1093/pcmedi/pbz028. Epub 2019 Dec 8. PMID: 31886034; PMCID: PMC6927097.
Precision Clinical Medicine 2019 Dec;2(4):221-234
Scientific Reports I December 2015
zhang Y, Zhong JF, Qiu H, MacLellan WR, Marbán E, Wang C (corresponding). Epigenomic Reprogramming of Adult Cardiomyocyte-Derived Cardiac Progenitor Cells. doi: 10.1038/srep17686. Erratum in: Sci Rep. 2017 Oct 20;7:46907. PMID: 26657817; PMCID: PMC4677315.
Scientific Reports 2015 Dec 14;5:17686
Nature Communications I May 2014
Yu Y, Fuscoe JC, Zhao C, Guo C, Jia M, Qing T, Bannon DI, Lancashire L, Bao W, Du T, Luo H, Su Z, Jones WD, Moland CL, Branham WS, Qian F, Ning B, Li Y, Hong H, Guo L, Mei N, Shi T, Wang KY, Wolfinger RD, Nikolsky Y, Walker SJ, Duerksen-Hughes P, Mason CE, Tong W, Thierry-Mieg J, Thierry-Mieg D, Shi L, Wang C (corresponding). A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. doi: 10.1038/ncomms4230. PMID: 24510058; PMCID: PMC3926002.
Nature Communications 2014;5:3230


Nature Biotechnology I September 2014
Wang C, Gong B, Bushel PR, Thierry-Mieg J, Thierry-Mieg D, Xu J, Fang H, Hong H, Shen J, Su Z, Meehan J, Li X, Yang L, Li H, Łabaj PP, Kreil DP, Megherbi D, Gaj S, Caiment F, van Delft J, Kleinjans J, Scherer A, Devanarayan V, Wang J, Yang Y, Qian HR, Lancashire LJ, Bessarabova M, Nikolsky Y, Furlanello C, Chierici M, Albanese D, Jurman G, Riccadonna S, Filosi M, Visintainer R, Zhang KK, Li J, Hsieh JH, Svoboda DL, Fuscoe JC, Deng Y, Shi L, Paules RS, Auerbach SS, Tong W. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. doi: 10.1038/nbt.3001. Epub 2014 Aug 24. PMID: 25150839; PMCID: PMC4243706.
Nature Biotechnology 2014 Sep;32(9):926-32


Nature Biotechnology i September 2014
SEQC/MAQC-III Consortium. Su Z, Labaj PP, Li S, Thierry-Mieg J, Thierry-Mieg D, Shi W, Wang C (project leader). et. al A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. doi: 10.1038/nbt.2957. Epub 2014 Aug 24. PMID: 25150838; PMCID: PMC4321899.
Nature Biotechnology 2014 Sep;32(9):903-14

Nature Biotechnology I September 2014
Li S, Łabaj PP, Zumbo P, Sykacek P, Shi W, Shi L, Phan J, Wu PY, Wang M, Wang C, Thierry-Mieg D, Thierry-Mieg J, Kreil DP, Mason CE. Detecting and correcting systematic variation in large-scale RNA sequencing data. doi: 10.1038/nbt.3000. Epub 2014 Aug 24. PMID: 25150837; PMCID: PMC4160374.
Nature Biotechnology 2014 Sep;32(9):888-95

Nature Communications I September 2014
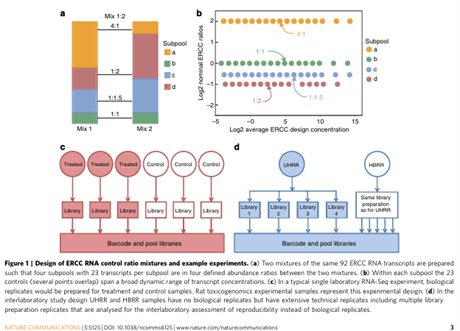
Munro SA, Lund SP, Pine PS, Binder H, Clevert DA, Conesa A, Dopazo J, Fasold M, Hochreiter S, Hong H, Jafari N, Kreil DP, Łabaj PP, Li S, Liao Y, Lin SM, Meehan J, Mason CE, Santoyo-Lopez J, Setterquist RA, Shi L, Shi W, Smyth GK, Stralis-Pavese N, Su Z, Tong W, Wang C, Wang J, Xu J, Ye Z, Yang Y, Yu Y, Salit M. Assessing technical performance in differential gene expression experiments with external spike-in RNA control ratio mixtures. doi: 10.1038/ncomms6125. PMID: 25254650.
Nature Communications 2014 Sep 25;5:5125